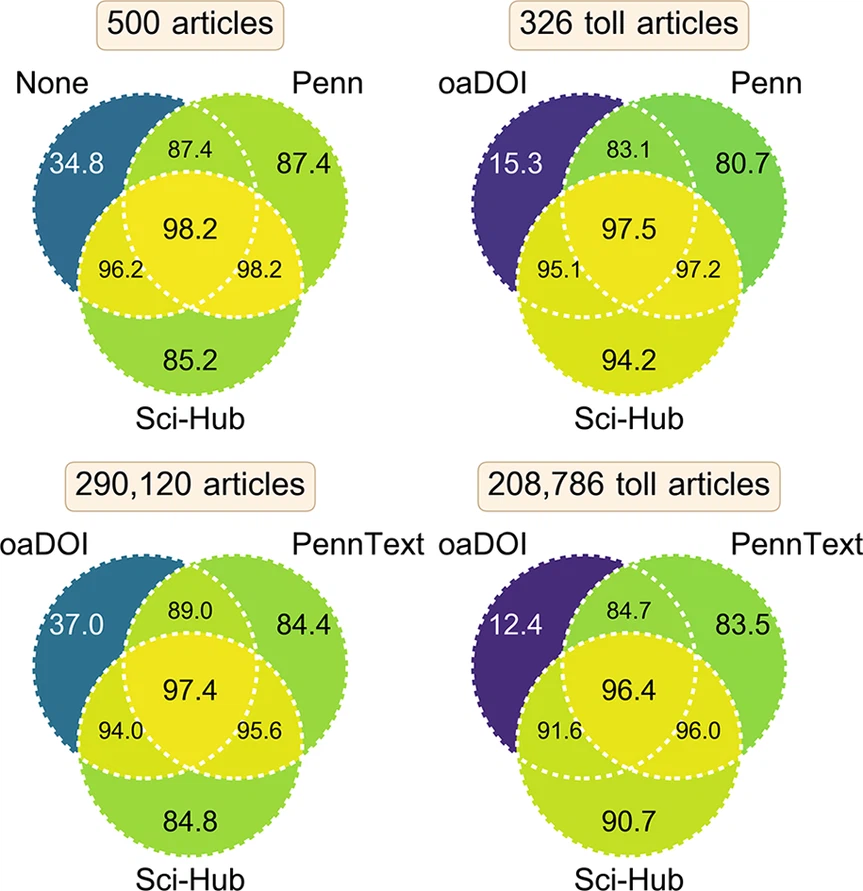
Testbed
Basic formatting
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur. Excepteur sint occaecat cupidatat non proident, sunt in culpa qui officia deserunt mollit anim id est laborum.
italic text
bold text
strike-through text
Text with extra blank lines above and below
- list item a
- list item b
- list item c
- ordered list item 1
- ordered list item 2
- ordered list item 3
- top level list item
- nested list item
- even deeper nested list item
- nested list item
Plain image:

Heading 1
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua.
Heading 2
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua.
Heading 3
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua.
Heading 4
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua.
Heading 5
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua.
Heading 6
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua.
| TABLE | Game 1 | Game 2 | Game 3 | Total |
|---|---|---|---|---|
| Anna | 144 | 123 | 218 | 485 |
| Bill | 90 | 175 | 120 | 385 |
| Cara | 102 | 214 | 233 | 549 |
It was the best of times it was the worst of times. It was the age of wisdom, it was the age of foolishness. It was the spring of hope, it was the winter of despair.
// some code with syntax highlighting
const popup = document.querySelector("#popup");
popup.style.width = "100%";
popup.innerText =
"Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua.";
This sentence has inline code, useful for making references to variables, packages, versions, etc. within a sentence.
Lorem ipsum dolor sit amet.
Consectetur adipiscing elit.
Sed do eiusmod tempor incididunt.
Hidden content
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua.
Jekyll Spaceship
| Stage | Direct Products | ATP Yields |
|---|---|---|
| Glycolysis
|
2 ATP | |
| 2 NADH | 3–5 ATP | |
| Pyruvaye oxidation | 2 NADH | 5 ATP |
| Citric acid cycle
|
2 ATP | |
| 6 NADH | 15 ATP | |
| 2 FADH | 3 ATP | |
| 30–32 ATP |
$ a * b = c ^ b $
$ 2^{\frac{n-1}{3}} $
$ \int_a^b f(x)\,dx. $
Components
Section
Section, background
Section, dark=true
Section, background dark=true
Section, size=wide
Section, size=full w/ figure

Figure



px width

% width

px height

px width, svg

% width, svg

px height, svg
Button
Icon
Lorem Ipsum Dolor
Feature
Title
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua.
List
List citations
2024
2023
2022
2021
2020
2019
2018
2017
2016
2015
2014
List projects
CAVE is a cloud-based platform that calculates and visualizes optimal metabolic pathways using flux balance analysis, and serves as a GEM quality control tool with network modification capabilities, all supported by a scalable serverless architecture.
Reaction Enzyme Mining & Evaluation (REME) Platform, the first integrated web platform for reaction enzyme mining and evaluation, is explicitly designed to solve two fundamental bioengineering problems: 1. Mining potential enzymes for natural and non-natural bio-related reactions; 2. Evaluating the catalysis possibility of provided (or mined) enzymes and reactions.
WPHP enables users to design optimal synthetic pathways for products in different host organisms. The results provide the yield of product, the flux distribution of pathway and the enzyme or genes encoding every reaction and the visual representation of any pathway.
QHEPath is a cloud-based, serverless web tool for quantitative heterologous pathway design. It relies on a quality-controlled universal model from the BiGG database and employs an algorithm to compute multiple suboptimal and optimal pathways for product synthesis.
ECRECer is a deep learning based bioinformatic toolkit to recommend EC numbers with high accuracy and high recall.
Model Quality Control (MQC) is an online tool designed for the quality assessment and automatic correction of genome-scale metabolic network models (GEMs). It focuses on identifying errors such as incorrect exchange reaction boundaries, faulty net generation of reducing equivalents, energy, and metabolites. Furthermore, MQC verifies whether the yield of each carbon-containing metabolite exceeds the theoretical optimal yield and evaluates if the organism can be simulated to grow while ensuring that the biomass composition equation is mass balanced. MQC then automatically corrects these errors, with a specific emphasis on ensuring accurate quantitative pathway calculations from the model.
OptME is a user-friendly web platform designed to assist researchers in metabolic engineering design. OptME simplifies computational target design—a key step in the DBTL (Design-Build-Test-Learn) cycle—by narrowing experimental scope and prioritizing metabolic engineering targets.OptME integrates classic algorithms like FSEOF (PMID:20348305) and OptForce (PMID:20419153) with newer approaches such as iBridge (PMID:37935194). It supports both stoichiometric-based and enzyme-constrained models (PMID:35858410), offering a comprehensive toolset that connects computational models with experimental workflows.
GEDpm-cg enables users to perform the efficient, automated and high-throughput design of single nucleotide editing in C. glutamicum chromosome. The counter-selection HR system and the overlap-based assembly method were chosen as the loading techniques. Homologous arms and primers required for genetic modification, vector DNA assembly and sequencing verification were provided as design results
AutoESD is a web tool for Automatic Editing Sequence Design for genetic manipulation of microorganisms, and it enables users to perform the precise, automated and high-throughput design of sequence manipulation tasks across species and technical variations, at any genomic locus for all manipulation types with adequate sequence length. The screening-marker-based homologous-recombination system and the overlap-based assembly method were chosen as the loading techniques. Homologous arms and primers required for sequence manipulation, vector DNA assembly and sequencing verification were provided as design results.
AutoESDCas is a A web tool for Automatic Editing Sequence Design for genetic manipulation of microorganisms based on CRISPR/Cas system, and it enables users to perform the precise, automated and high-throughput design of genome editing tasks. It facilitates all types of genetic manipulation as well as different CRISPR/Cas system-mediated homologous recombination genome editing technology variants and experimental requirements. The editing sequences such as sgRNAs, homology arms and primers for the whole-workflow of genome editing can be designed. Based on these editing sequences, genome editing experiments could be implemented.
FSsgRNA-Analyzer is A cloud platform for NGS data processing in CRISPR based functional screening, and it enables experimental biologist to perform the sgRNA data analysis easily during CRISPR based functional screening. The cloud platform is based on a novel serverless architecture, which enables high reliability, robustness and scalability. After users uploading raw sequence data and target sgRNA data, FSsgRNA-Analyzer can execute the primary analysis for NGS data (read cleaning, mapping and sgRNA counting), then perform the secondary analysis (correlation or differential analysis) among samples under different culture/selection conditions.
GSCAS-MTM is a web tool designed to generate optimized schedule trees for the parallel construction of a large number of objective strains. Our tool utilizes two complementary algorithms, GSCAS (Greedy Search of Common Ancestor Strains) and MTM (Minimizing Total Manipulations), to efficiently design strain construction schedules.
DNA synthesis is widely used in synthetic biology to construct and assemble de novo nucleotides, such as synthetic engineered protein, artificial synthetic genome. Many sequence features (e.g., GC content, repeats) are known to affect the process of DNA synthesis. SCP4SSD, a serverless cloud platform, was developed for nucleotide sequence synthesis difficulty prediction.
AutoVTC is a web tool specifically designed to streamline comprehensive, automated, and high-throughput mutation scanning for targeted genetic manipulation. This efficient tool excels in calling high-quality single-nucleotide polymorphisms (SNPs), insertions/deletions (InDels), and double peaks from Sanger sequencing chromatograms.
List team members
List blog posts
2023

Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua.
2021
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua.
2019
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua.
Citation
Card
Portrait
Post Excerpt
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua.
Alert
Tip Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua.
Help Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua.
Info Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua.
Success Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua.
Warning Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua.
Error Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua.
Tags
Float
Figures
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur. Excepteur sint occaecat cupidatat non proident, sunt in culpa qui officia deserunt mollit anim id est laborum.
Code
const test = "Lorem ipsum dolor sit amet, consectetur adipiscing elit.";
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Nulla facilisi etiam dignissim diam quis. Id aliquet lectus proin nibh nisl condimentum id venenatis a. Tristique magna sit amet purus gravida quis blandit turpis cursus. Ultrices eros in cursus turpis massa tincidunt dui ut ornare. A cras semper auctor neque vitae tempus quam pellentesque nec. At tellus at urna condimentum mattis pellentesque. Ipsum consequat nisl vel pretium. Ultrices mi tempus imperdiet nulla malesuada pellentesque elit eget gravida. Integer vitae justo eget magna fermentum iaculis eu non diam. Mus mauris vitae ultricies leo integer malesuada nunc vel. Leo integer malesuada nunc vel risus. Ornare arcu odio ut sem nulla pharetra. Purus semper eget duis at tellus at urna condimentum. Enim neque volutpat ac tincidunt vitae semper quis lectus.
Grid
Regular
With Markdown images

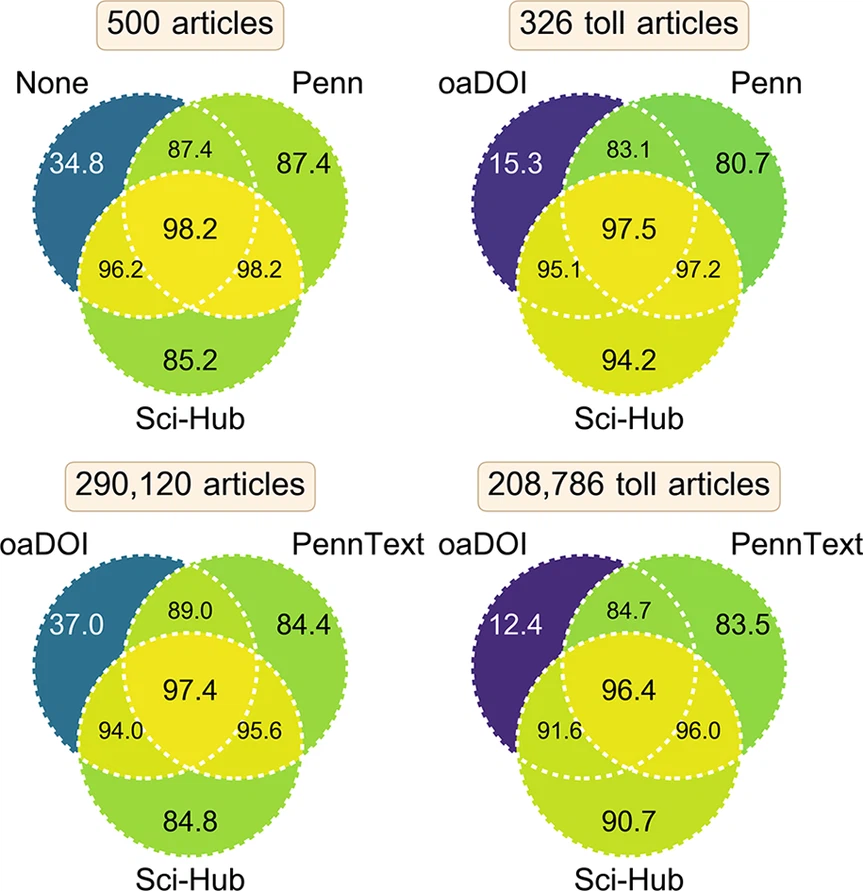
![]()
![]()
![]()
Square
With figure components
Grid of citations
2024
2023
2022
2021
2020
2019
2018
2017
2016
2015
2014
Grid of blog posts
2023

Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua.
2021
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua.
2019
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua.
Cols
Text
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua.
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Nulla facilisi etiam dignissim diam quis. Id aliquet lectus proin nibh nisl condimentum id venenatis a.
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Nulla facilisi etiam dignissim diam quis. Id aliquet lectus proin nibh nisl condimentum id venenatis a. Tristique magna sit amet purus gravida quis blandit turpis cursus. Ultrices eros in cursus turpis massa tincidunt dui ut ornare. A cras semper auctor neque vitae tempus quam pellentesque nec. At tellus at urna condimentum mattis pellentesque. Ipsum consequat nisl vel pretium. Ultrices mi tempus imperdiet nulla malesuada pellentesque elit eget gravida. Integer vitae justo eget magna fermentum iaculis eu non diam. Mus mauris vitae ultricies leo integer malesuada nunc vel. Leo integer malesuada nunc vel risus. Ornare arcu odio ut sem nulla pharetra. Purus semper eget duis at tellus at urna condimentum. Enim neque volutpat ac tincidunt vitae semper quis lectus.
Images

Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua.
Code
const test = "Lorem ipsum dolor sit amet";
const test = "Lorem ipsum dolor sit amet";
const test = "Lorem ipsum dolor sit amet";